Analyses
Here are listed the single-cell RNA Sequencing (scRNA-Seq) data analyses I made. The raw data and/or count matrices are available under Data. The code to make the analyses from the count matrices up to the manuscript is available under Code. The code development is tracked with GitHub but the analyses associated with the manuscript are made identifiable using Zenodo. To enable you to explore the analysis, you can access the GitHub Pages under Analysis. The index of the GitHub Pages has been built using OrganizeHTML (more information in Tools). If you have any questions, feel free to open an issue in the associated Github repository.
|
Reanalysis of a public dataset of hair follicle cells in physiological condition |
scRNA-Seq atlas of hair follicle cells in the context of Hidradenitis suppurativa |
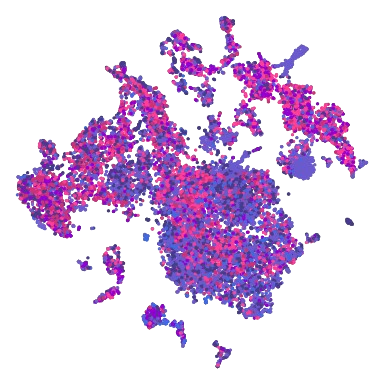 Data | Code | Analysis
Data | Code | Analysis
Onfroy et al. (2025), DOI: 10.1145/3736731.3746138 |
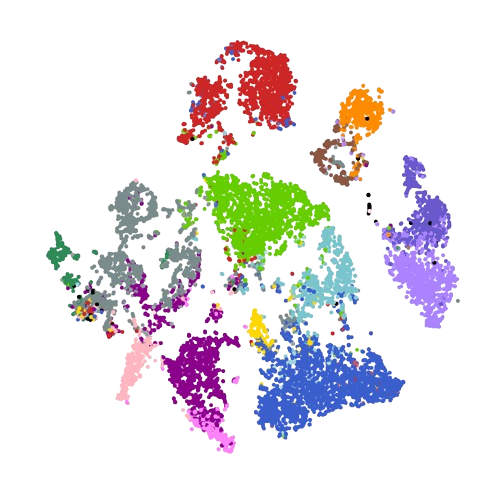 Data | Code | Analysis
Data | Code | Analysis
Onfroy et al. (2025), DOI: 10.1101/2025.05.28.656362 |
|
scRNA-Seq atlas of NF1-associated Peripheral Nerve Sheath Tumors (mouse) |
scRNA-Seq atlas of NF1-associated Peripheral Nerve Sheath Tumors (patients) |
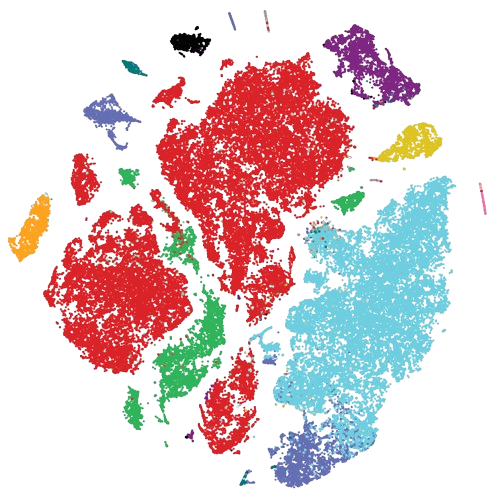 Data | Code | Analysis
Data | Code | Analysis
Radomska et al. (2025), DOI: In Press |
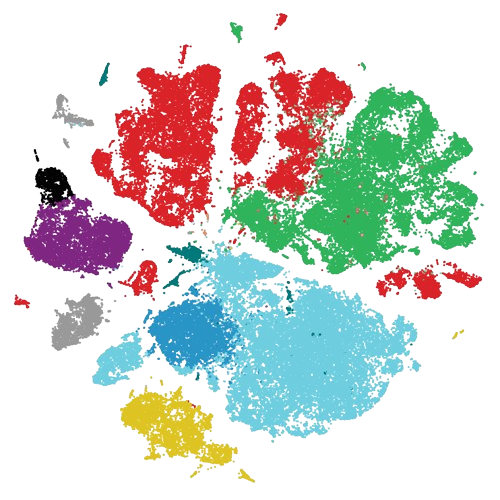 Data | Code | Analysis
Data | Code | Analysis
Radomska et al. (2025), DOI: In Press |
|
scRNA-Seq atlas of subcutaneous nerves in the contexte of NF1 (mouse) |
... |
 Data | Code | Analysis
Data | Code | Analysis
Mansour, Onfroy et al. (2025), DOI: 10.1101/2025.07.04.662123 |
... |